Many quantitative science fields are adopting the paradigm of predictive modeling
using machine learning. We welcome this development. At the same time, as researchers whose interests include
the strengths and limits of machine learning, we have concerns about reproducibility and overoptimism.
There are many reasons for caution:
We focus on reproducibility issues in ML-based science, which involves making a scientific claim using the performance of the ML model as evidence. There is a much better known reproducibility crisis in research that uses traditional statistical methods. We also situate our work in contrast to other ML domains, such as methods research (creating and improving widely-applicable ML methods), ethics research (studying the ethical implications of ML methods), engineering applications (building or improving a product or service), and modeling contests (improving predictive performance on a fixed dataset created by an independent third party). Investigating the validity of claims in all of these areas is important, and there is ongoing work to address reproducibility issues in these domains.
A non-exhaustive categorization of focus areas in ML literature. In our work, we focus on ML-based science.
The running list below consists of papers that highlight reproducibility failures or pitfalls in ML-based science. We find 41 papers from 30 fields where errors have been found, collectively affecting 648 papers and in some cases leading to wildly overoptimistic conclusions. In each case, data leakage causes errors in the modeling process. (Table updated in May 2024)
| Field | Paper | Year | Num. papers reviewed | Num. papers w/pitfalls | Pitfalls |
|---|---|---|---|---|---|
| Family relations | Heyman et al. | 2001 | 15 | 15 | No train-test split |
| Medicine | Bouwmeester et al. | 2012 | 71 | 27 | No train-test split |
| Molecular biology | Park et al. | 2012 | 59 | 42 | Non-independence |
| Neuroimaging | Whelan et al. | 2014 | — | 14 | No train-test split; Feature selection on train and test set |
| Bioinformatics | Blagus et al. | 2015 | — | 6 | Pre-processing on train and test sets together |
| Autism Diagnostics | Bone et al. | 2015 | — | 3 | Duplicates across train-test split; Sampling bias |
| Nutrition research | Ivanescu et al. | 2016 | — | 4 | No train-test split |
| Software engineering | Tu et al. | 2018 | 58 | 11 | Temporal leakage |
| Toxicology | Alves et al. | 2019 | — | 1 | Duplicates across train-test split |
| Clinical epidemiology | Christodoulou et al. | 2019 | 71 | 48 | Feature selection on train and test set |
| Satelitte imaging | Nalepa et al. | 2019 | 17 | 17 | Non-independence between train and test sets |
| Tractography | Poulin et al. | 2019 | 4 | 2 | No train-test split |
| Brain-computer interfaces | Nakanishi et al. | 2020 | — | 1 | No train-test split |
| Histopathology | Oner et al. | 2020 | — | 1 | Non independence between train and test sets |
| Neuropsychiatry | Poldrack et al. | 2020 | 100 | 53 | No train-test split; pre-processing on train and test sets together |
| Molecular biology | Urban et al. | 2020 | — | 12 | Non independence between train and test sets |
| Neuroimaging | Ahmed et al. | 2021 | — | 1 | Non independence between train and test sets |
| Neuroimaging | Li et al. | 2021 | 122 | 18 | Non independence between train and test sets |
| IT Operations | Lyu et al. | 2021 | 9 | 3 | Temporal leakage |
| Medicine | Filho et al. | 2021 | — | 1 | Illegitimate features |
| Radiology | Roberts et al. | 2021 | 62 | 16 | No train-test split; duplicates in train and test sets; sampling bias |
| Information retrieval | Berrendorf et al. | 2021 | — | 3 | No train-test split; Feature selection on training and test sets |
| Neuropsychiatry | Shim et al. | 2021 | — | 1 | Feature selection on training and test sets |
| Medicine | Vandewiele et al. | 2021 | 24 | 21 | Feature selection on train-test sets; Non-independence between train and test sets; Sampling bias |
| Computer security | Arp et al. | 2022 | 30 | 22 | No train-test split; Pre-processing on train and test sets together; Illegitimate features; others |
| Genomics | Barnett et al. | 2022 | 41 | 23 | Feature selection on training and test sets |
| Molecular biology | Li et al. | 2022 | 18 | 13 | Non independence between training and test sets |
| Genetics | Whalen et al. | 2022 | — | 11 | Pre-processing on train and test sets; Non independence between training and test sets; Sampling bias |
| Radiology | Gidwani et al. | 2022 | 50 | 39 | No train-test split; Pre-processing on train and test sets; Feature selection on training and test sets; Illegitimate features; Sampling bias |
| Sustainable energy | Geslin et al. | 2023 | — | 1 | Illegitimate features |
| Medicine | Ren et al. | 2023 | — | 5 | Pre-processing on training and test sets; Illegitimate features |
| Medicine | Akay et al. | 2023 | 65 | 27 | No train-test split |
| Pharmaceutical sciences | Murray et al. | 2023 | — | 2 | Pre-processing on training and test sets; Feature selection on training and train test sets |
| Political Science | Kapoor and Narayanan | 2023 | 12 | 4 | Pre-processing on training and test sets; Illegitimate features; Temporal leakage |
| Ecology | Stock et al. | 2023 | — | 3 | Sampling bias |
| Computer networks | Kostas et al. | 2023 | — | 4 | Non-independence between training and test sets |
| Law | Medvedeva et al. | 2023 | 171 | 156 | Illegitimate features; Temporal leakage; Non independence between training and test sets |
| Dermatology | Abhishek et al. | 2024 | — | 2 | No train-test split; Duplicates across train-test split |
| Medicine | Kwong et al. | 2024 | 15 | 3 | No train-test split |
| Mining | Munagala et al. | 2024 | — | 4 | Pre-processing on training and test sets |
| Affective computing | Varanka et al. | 2024 | — | 8 | Pre-processing on training and test sets; Feature selection on training and test sets |
Data leakage has long been recognized as a leading cause of errors in ML applications. In formative work on leakage, Kaufman et al. provide an overview of different types of errors and give several recommendations for mitigating these errors. Since this paper was published, the ML community has investigated the impact of leakage in several engineering applications and modeling competitions. However, leakage occurring in ML-based science has not been comprehensively investigated. As a result, mitigations for data leakage in scientific applications of ML remain understudied.
Our taxonomy of data leakage highlights several failure modes which are prevalent in ML-based science. To
address leakage, researchers using ML methods need to connect the performance of their ML models to their
scientific claims. To detect cases of leakage, we provide a template for a model info sheet which should be included when
making a scientific claim using predictive modeling. The template consists of precise arguments needed to
justify the absence of leakage, and is inspired by Mitchell et
al.'s model cards for increasing the transparency of ML models.
Model info sheets can be voluntarily used by researchers to detect leakage. Of course, model info sheets can’t
prevent researchers from making false claims, but we hope they can make errors more apparent. Note that for
model info sheets to be verified, the analysis must be computationally reproducible. Also, model info sheets
don’t address reproducibility issues other than leakage.
We find that prominent studies on civil war prediction claiming superior performance of ML models over baseline Logistic Regression models fail to reproduce. Our results provide two reasons to be skeptical of the use of ML methods in this research area, by both questioning their usefulness and highlighting the pitfalls of applying them correctly. While none of these errors could have been caught by reading the papers, our model info sheets enable the detection of leakage in each case.
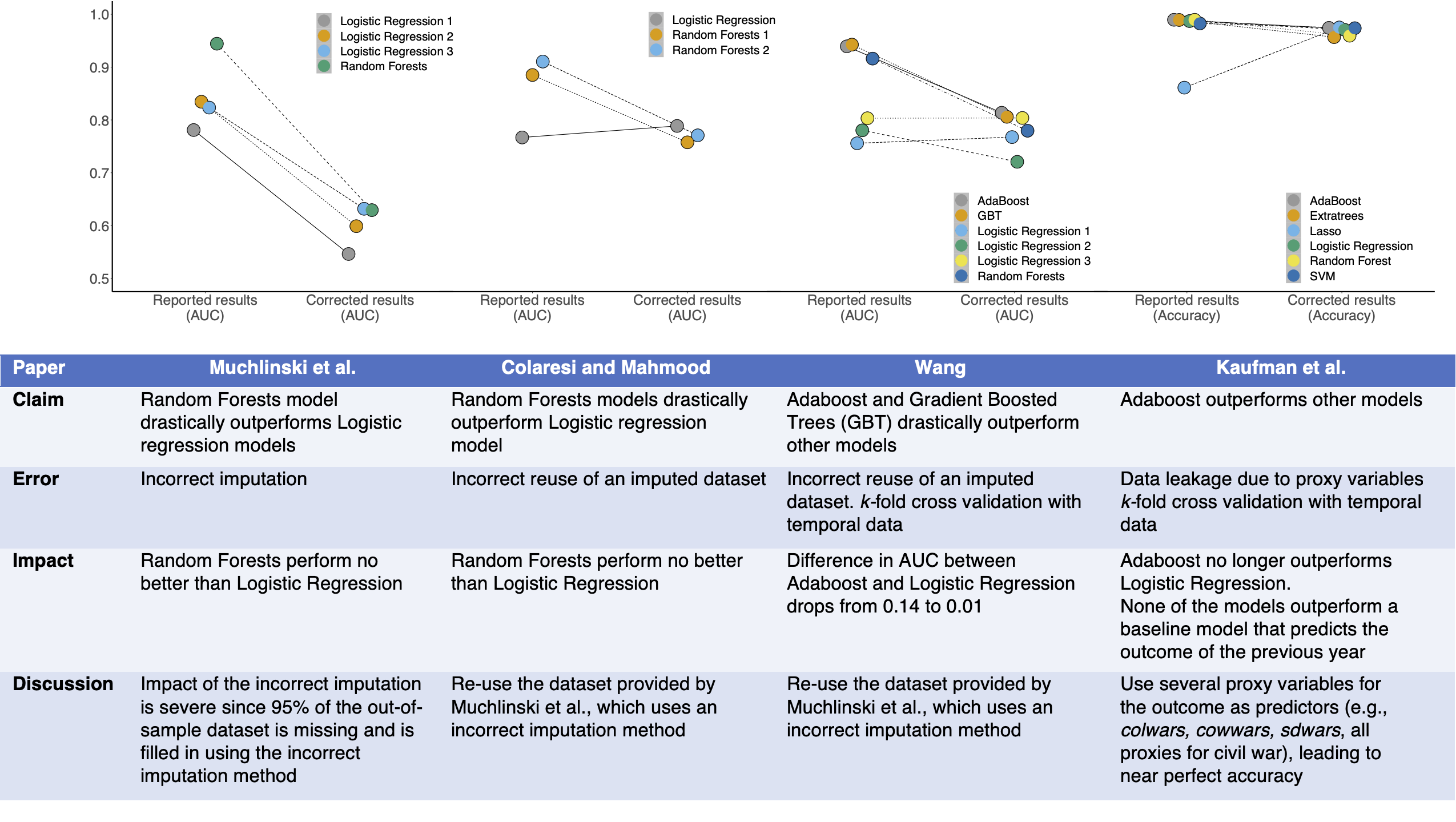
A comparison of reported and corrected results in civil war prediction papers published in top Political Science journals. The main findings of each of these papers are invalid due to various forms of data leakage: Muchlinski et al. impute the training and test data together, Colaresi & Mahmood and Wang incorrectly reuse an imputed dataset, and Kaufman et al. use proxies for the target variable which causes data leakage. The use of model info sheets would detect leakage in every paper. When we correct these errors, complex ML models (such as Adaboost and Random Forests) do not perform substantively better than decades-old Logistic Regression models for civil war prediction in each case. Each column in the table outlines the impact of leakage on the results of a paper.
We acknowledge that there isn't consensus about the term reproducibility, and there have been a number of
recent attempts to define the term and create consensus. One possible definition is computational
reproducibility — when the results in a paper can be replicated using the exact code and dataset provided by
the authors. We argue that this definition is too narrow because even cases of outright bugs in the code would
not be considered irreproducible under this definition. Therefore we advocate for a standard where bugs and
other errors in data analysis that change or challenge a paper's findings constitute irreproducibility. We
elaborate this perspective here.
Reproducibility failures don’t mean a claim is wrong, just that evidence presented falls short of the accepted
standard or that the claim only holds in a narrower set of circumstances than asserted. We don’t view
reproducibility failures as signs that individual authors or teams are careless, and we don’t think any
researcher is immune. One of us (Narayanan) has had multiple such failures in his applied-ML work and expects
that it will probably happen again.
We call it a crisis for two related reasons. First, reproducibility failures in ML-based science are systemic.
In nearly every scientific field that has carried out a systematic study of reproducibility issues, papers are
plagued by common pitfalls. In many systematic reviews, a majority of the papers reviewed suffer from these
pitfalls. Second, despite the urgency of addressing reproducibility failures, there aren’t yet any systemic
solutions.